This vignette describes the two implemented methods for blockmodeling in signed networks.
Traditional Blockmodeling
In signed blockmodeling, the goal is to determine k
blocks of nodes such that all intra-block edges are positive and
inter-block edges are negative. In the example below, we construct a
network with a perfect block structure with
sample_islands_signed(). The network consists of 10 blocks
with 10 vertices each, where each block has a density of 1 (of positive
edges). The function signed_blockmodel() is used to
construct the blockmodel. The parameter k is the number of
desired blocks. alpha is a trade-off parameter. The
function minimizes
,
where
is the total number of negative ties within blocks and
be the total number of positive ties between blocks.
g <- sample_islands_signed(10,10,1,20)
clu <- signed_blockmodel(g,k = 10,alpha = 0.5)
table(clu$membership)
#>
#> 1 2 3 4 5 6 7 8 9 10
#> 10 10 10 10 10 10 10 10 10 10
clu$criterion
#> [1] 0The function returns a list with two entries. The block membership of nodes and the value of .
The function ggblock() can be used to plot the outcome
of the blockmodel (ggplot2 is required).
ggblock(g,clu$membership,show_blocks = TRUE)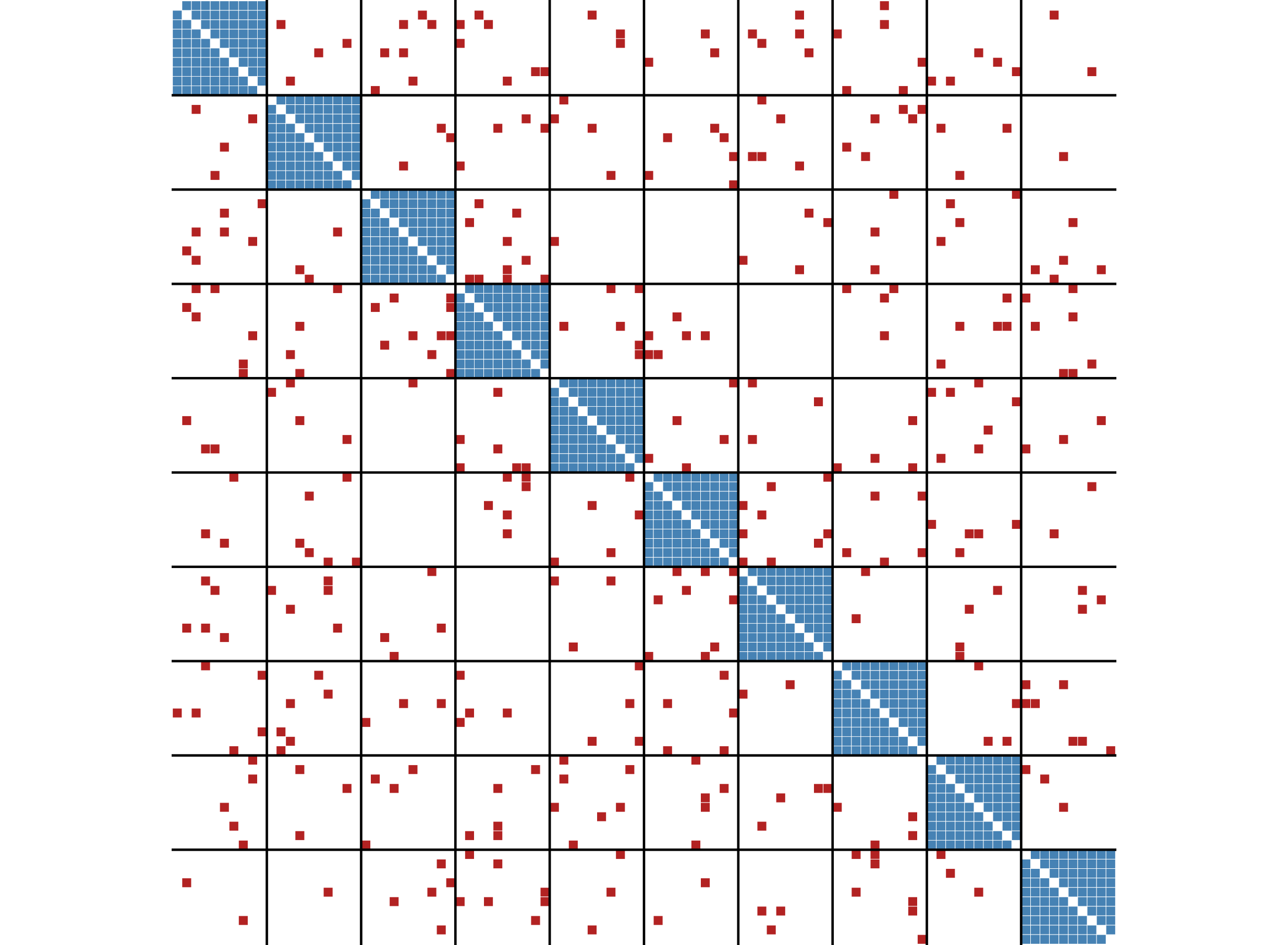
If the parameter annealing is set to TRUE, simulated
annealing is used in the optimization step. This generally leads to
better results but longer runtimes.
data("tribes")
set.seed(44) #for reproducibility
signed_blockmodel(tribes,k = 3,alpha=0.5,annealing = TRUE)
#> $membership
#> [1] 1 1 2 2 3 2 2 2 3 3 2 2 3 3 1 1
#>
#> $criterion
#> [1] 2
signed_blockmodel(tribes,k = 3,alpha=0.5,annealing = FALSE)
#> $membership
#> [1] 1 1 2 2 3 2 2 2 3 3 2 2 3 3 1 1
#>
#> $criterion
#> [1] 2Generalized Blockmodeling
The function signed_blockmodel() is only able to provide
a blockmodel where the diagonal blocks are positive and off-diagonal
blocks are negative. The function
signed_blockmodel_general() can be used to specify
different block structures. In the below example, we construct a network
that contains three blocks. Two have positive and one has negative
intra-group ties. The inter-group edges are negative between group one
and two, and one and three. Between group two and three, all edges are
positive.
g1 <- g2 <- g3 <- make_full_graph(5)
V(g1)$name <- as.character(1:5)
V(g2)$name <- as.character(6:10)
V(g3)$name <- as.character(11:15)
g <- Reduce("%u%",list(g1,g2,g3))
E(g)$sign <- 1
E(g)$sign[1:10] <- -1
g <- add.edges(g,c(rbind(1:5,6:10)),attr = list(sign=-1))
g <- add.edges(g,c(rbind(1:5,11:15)),attr = list(sign=-1))
g <- add.edges(g,c(rbind(11:15,6:10)),attr = list(sign=1))The parameter blockmat is used to specify the desired
block structure.
set.seed(424) #for reproducibility
blockmat <- matrix(c(1,-1,-1,-1,1,1,-1,1,-1),3,3,byrow = TRUE)
blockmat
#> [,1] [,2] [,3]
#> [1,] 1 -1 -1
#> [2,] -1 1 1
#> [3,] -1 1 -1
general <- signed_blockmodel_general(g,blockmat,alpha = 0.5)
traditional <- signed_blockmodel(g,k = 3,alpha = 0.5,annealing = TRUE)
c(general$criterion,traditional$criterion)
#> [1] 0 6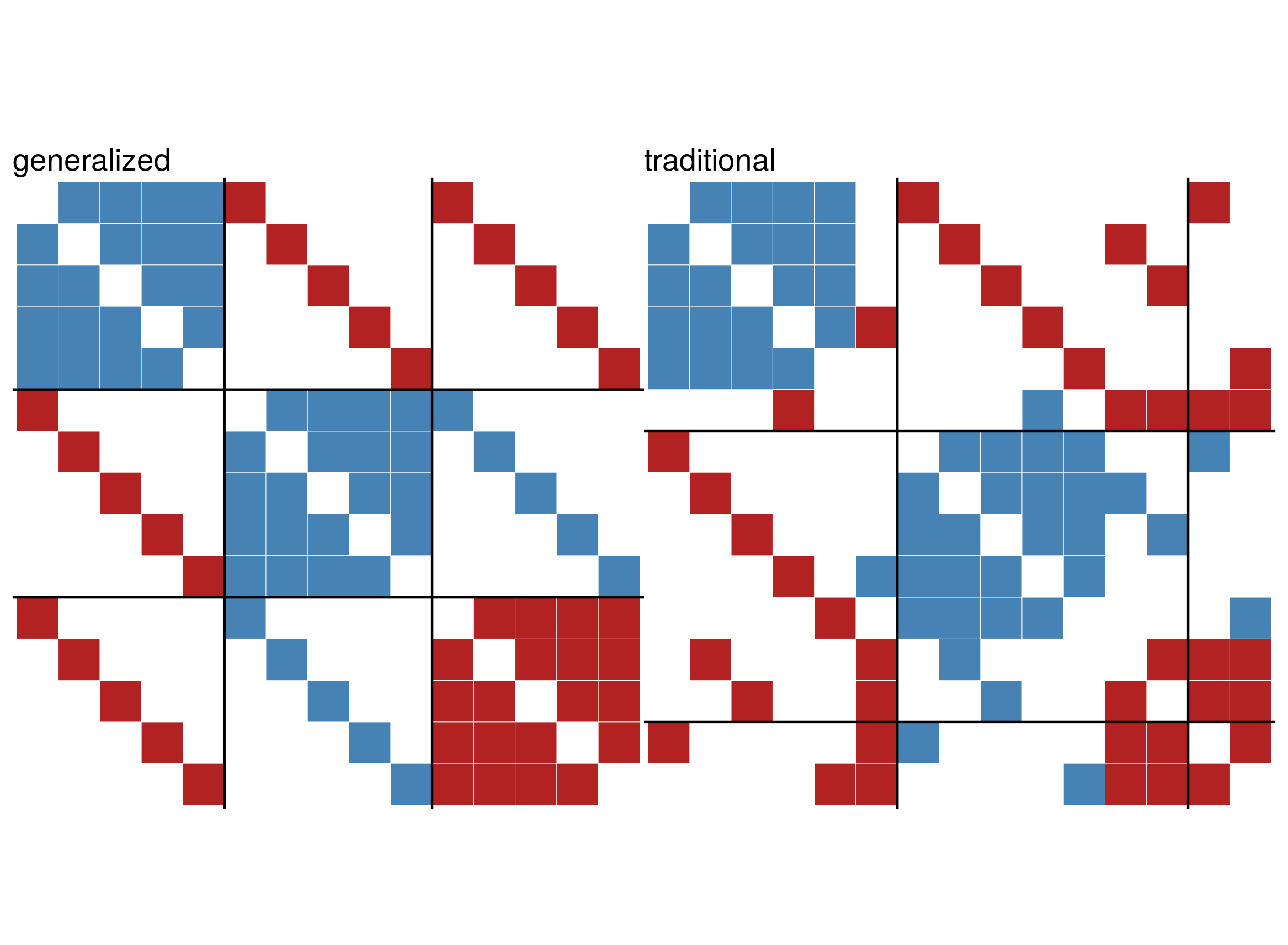
References
Doreian, Patrick, and Andrej Mrvar. 1996. “A Partitioning Approach to Structural Balance.” Social Networks 18 (2): 149–68.
Doreian, Patrick, and Andrej Mrvar. 2009. “Partitioning Signed Social Networks.” Social Networks 31 (1): 1–11.
Doreian, Patrick, and Andrej Mrvar. 2015. “Structural Balance and Signed International Relations.” Journal of Social Structure 16: 1.